T3/Breedbase Workshop
 September 8th - 9th, 2020
September 8th - 9th, 2020
 David Waring: djw64@cornell.edu
David Waring: djw64@cornell.edu
Important Links
T3/Breedbase Production Databases:
- T3/Wheat - https://wheat.triticeaetoolbox.org
- T3/Barley - https://barley.triticeaetoolbox.org
- T3/Oat - https://oat.triticeaetoolbox.org
T3/Breedbase Sandbox Databases:
- T3/Wheat [Sandbox] - https://wheat-sandbox.triticeaetoolbox.org
- T3/Barley [Sandbox] - https://barley-sandbox.triticeaetoolbox.org
- T3/Oat [Sandbox] - https://oat-sandbox.triticeaetoolbox.org
Phenotype Upload Instructions:
- Wheat - https://wheat-sandbox.triticeaetoolbox.org/help/phenotype_upload_workflow
- Barley - https://barley-sandbox.triticeaetoolbox.org/help/phenotype_upload_workflow
- Oat - https://oat-sandbox.triticeaetoolbox.org/help/phenotype_upload_workflow
Germplasm Search Tool:
- Wheat - https://synonyms.triticeaetoolbox.org/?db=T3/Wheat
- Barley - https://synonyms.triticeaetoolbox.org/?db=T3/Barley
- Oat - https://synonyms.triticeaetoolbox.org/?db=T3/Oat
Video Recordings:
- Introduction - https://www.youtube.com/watch?v=8J6cZtSgxU4
- Locations - https://www.youtube.com/watch?v=Q-EfBXNQ1JI
- Accessions - https://www.youtube.com/watch?v=i5BZvS8X_fc
- Trials - https://www.youtube.com/watch?v=OfdpGa9EfBU
- Traits - https://www.youtube.com/watch?v=yU3P6fZM_YE
- Phenotype Observations - https://www.youtube.com/watch?v=W3PVJLyUOkQ
- Adopting Breedbase (Jessica Rutkoski) - https://www.youtube.com/watch?v=1X4Q3RqLB6o
Blank Upload Templates
 Location Template
Location Template
 Accession Template
Accession Template
 Trial Template (trial information and plot layout)
Trial Template (trial information and plot layout)
 Phenotype Template (trait observations)
Phenotype Template (trait observations)
Trait Lookup Tables
Day One
What is T3? 
T3 is The Triticeae Toolbox:
- a centralized database for small grains breeders (wheat, oat, barley)
- combines accession information, phenotype trial data, and genotype data from various sources
- allows users to create custom datasets
- provides summary and analytical tools
Overview of T3/Breedbase
What is Happening?
- T3 is changing the database and website used for collecting, analyzing and displaying breeding data
- Existing data has been transferred to the new database
- New data will be added to the new database - the old database will no longer be receiving new data
- The old database will remain accessible as a data archive
Current Status
- Wheat: transition complete
- Oat: transition complete
- Barley: transition complete, some organization still needed
What is Breedbase?
- a “comprehensive breeding management and analysis software”
- Database, analytical tools and website
- Developed by Lukas Mueller’s lab at Boyce Thompson Institute
- Currently used by multiple crops: cassava, sweet potato, banana, rice, Solanaceae crops, others
Why is T3 transitioning?
- more developers
- core development team at BTI
- already being used by multiple crops
- unified database
- sharing of ideas, database structures, tools, etc across breeding programs
- unique features
- trial design
- seedlot management
- barcodes
Workshop Goals
Demonstrate how to upload field trials
- Demonstrate the entire process of getting a field trial into the database
- checking for existing Accessions
- updating existing & adding new Accessions
- creating an existing Trial
- working with breedbase traits
- adding phenotype observations
Demonstrate Breedbase integration into a breeding program
- Jessica will demonstrate how she uses breedbase in the University of Illinois wheat breeding program
Try uploading your own data
- Participants will have the opportunity to try uploading a dataset of their own
- We will be available to help and troubleshoot any issues that arise
Wrap-Up Discussion | Day Two
- Discuss problems that were encountered
- show solutions for them
- discuss how we might make changes to fix the problem
- Introduce some analysis tools that are included with breedbase
- discuss if these may be useful
- allow us to prioritize in setting them up
Phenotype Submission Workflow
Database Organization
Each crop has a separate production and sandbox database.
- The production database:
- contains curated data and is read-only.
- Data cannot be added directly to the production database by users.
- This is the database you should go to when you want to access data for research purposes.
- The sandbox database:
- contains a copy of the production database (typically copied once a week).
- Data can be added directly to the sandbox database and modified by users.
- This is the database you should use when you want to test-load data.
Submission Overview
Two ways to submit data:
- Using the Sandbox
 this is the method we’ll be using for the workshop
this is the method we’ll be using for the workshop
- how it works:
- the user creates the necessary upload templates (new accessions, trials, observations, etc)
- the user uploads all of the data to the sandbox
- the user can verify the trials to make sure everything looks correct
- the user submits each trial to the production database
- the curator approves the submission
- the trial is added to the production database
- this method is good for:
- uploading a small number of trials (such as a single year’s worth of trials)
- testing a dataset (you want to see how the data looks in the database)
- familiarizing yourself with the database (you can upload data without changing the production database)
- how it works:
- Submitting Templates
- how it works:
- the user creates the necessary upload templates (new accessions, trials, observations, etc)
- the user sends the templates to the curator (via email or the submission form)
- the curator approves the submission
- the trials are added to the production database
- this method is good for:
- submitting a large number of trials (such as a historical dataset)
- quickly submitting trial data (when you’re already familiar with the upload template format)
- how it works:
Instructions
Detailed upload instructions are available on the sandbox database for each crop:
Demo Data
Below is an excerpt from and a link to the demo trial data I’ll be adding to the database:
| Plot | Line | Row | Column | Rep | Block | grain yield (kg/ha) | test weight (g/L) | FHB Incidence (%) | FHB Severity (%) | DON Level (ppm) |
|---|---|---|---|---|---|---|---|---|---|---|
| 101 | VA14W-99 | 1 | 1 | 1 | 1 | 1984.58 | 739.01 | 53.15 | 8.91 | 1.13 |
| 102 | NC14-23372 | 1 | 2 | 1 | 1 | 2044.19 | 756.15 | 35.59 | 8.08 | 1.75 |
| 103 | VA13W-38 | 1 | 3 | 1 | 1 | 2356.95 | 787.56 | 36.02 | 20.78 | 1.52 |
| 104 | NC15-25672 | 1 | 4 | 1 | 1 | 2061.00 | 752.54 | 69.92 | 20.64 | 2.25 |
| 105 | NC14-23373 | 1 | 5 | 1 | 1 | 1535.94 | 769.60 | 78.59 | 9.40 | 1.38 |
The demo data contains observations from two locations: Ithaca, NY and Geneva, NY. There are ten lines sampled in three reps at each location with five traits observed: grain yield, test weight, FHB incidence, FHB severity, and DON level.
Create an Account
Each crop has a separate production and sandbox database. These are separate websites and copies of the database and as such have separate account management systems. You’ll need to create an account on each database you want access to.
You can create an account by clicking the New User button in the top-right of the toolbar. After filling out the registration form you should receive an account confirmation email from noreply@graingenes.org (check your Junk/Spam folder if you don’t see it in your Inbox) with a link to confirm your account. Once your account has been confirmed you can log in to the website.
Accounts created on a sandbox database will be automatically granted ‘submitter’ privileges. Accounts created on a production database will have read-only access.
Locations 
Use Case: You have trials that were located in two different towns. Are these locations already in the database? How do you add a new location?
View Existing Locations
You can view the locations that are already in the database by going to the Manage > Locations page. Here you’ll see a table and a map of all existing locations to search for your trial locations.
Add New Locations
If the location of your trial is not already in the database, there are two ways to add new locations from the Manage > Locations page:
- Using the Map
- Click the location of your trial on the correct spot on the map
- Click the Add Location link
- Fill out the form with the location details
- Uploading a Template
- Create an Excel file with the appropriate headers
- Fill in your location details
- Upload the form
Location Template
A location template includes the following information:
| Name | Abbreviation | Country Code | Country Name | Program | Type | Latitude | Longitude | Altitude | NOAA Station ID |
|---|---|---|---|---|---|---|---|---|---|
| Geneva, NY | GENNY | USA | United States of America | Cornell University | Farm | 42.880173 | -77.009083 | 187 | GHCND:USC00303177 |
- Name: The Town and State of the location.
- Abbreviation: A code used to identify the location. By convention, T3 uses the first 3 letters of the town and the 2 letter state abbreviation.
- Country Code: A standardized 3 letter country code.
- Country Name: The full name of the Country of the location.
- Program: The Breeding Program(s) that use the location. The Breeding Program must already exist in the database.
- Type: The location type (one of: Town, Farm, Field, Greenhouse, Screenhouse, Lab, Storage, Other).
- Latitude: The location latitude (in decimal degrees).
- Longitude: The location longitude (in decimal degrees) with locations in the Western Hemisphere having a negative number.
- Altitude: The altitude/elevation of the location (in meters).
- NOAA Station ID: The identifier of the closest NOAA weather station (or ‘none’ if one is not nearby).
 If you’re using R to extract your data to generate the breedbase templates, you can use the breedbase R package to query an API to get the lat, lon, altitude and NOAA Station ID of locations based on street addresses.
If you’re using R to extract your data to generate the breedbase templates, you can use the breedbase R package to query an API to get the lat, lon, altitude and NOAA Station ID of locations based on street addresses.
Accessions 
Use Case: You have a list of lines that were phenotyped in your trials. Are these lines already in the database? How do you update existing and add new lines?
Germplasm Search Tool
The Germplasm Search Tool is a separate tool that queries the breedbase database for germplasm lines and matches your line names up with existing database entries using a number of search methods. It is available under the Search menu as Bulk Accession Search. Links for each crop are:
The Germplasm Search Tool can be used to check the experimental list of lines for existing matches in the database and identify lines that:
- already exist in the database - line name matches an existing line
- exist under a different name - line name matches a synonym of an existing line or is similar to an existing line name and/or synonym
- need to be added to the database - there is no entry that is the same or similar to an existing name and/or synonym
The ten lines in the demo data are named:
BESS
ERNIE
LES15-5499
LES15-5605
NC14-23372
NC14-23373
NC15-25672
VA13W-174
VA13W-38
VA14W-99
There are two lines that will need to be added as Accessions in the database:
NC15-25672
VA14W-99
Accession Template
The Accession upload template contains properties about the germplasm lines being added to the database. If an entry exists with the same name in the database, the properties will be updated with the ones in the template. At a minimum, the accesion_name and species_name properties are required.
An Accession template can include the following information:
| accession_name | species_name | population_name | organization_name(s) | synonym(s) | variety(s) | country_of_origin(s) | notes(s) | accession_number(s) | purdy_pedigree | filial_generation |
|---|---|---|---|---|---|---|---|---|---|---|
| NC15-25672 | Triticum aestivum | North Carolina State University | NC15_25672 | PI 1234 | NC15-1/NC15-2 | 9 | ||||
| VA14W-99 | Triticum aestivum | Virginia Tech | VA14W 99 | CItr 5678 | VA14W-1/VA14W-2 | 9 |
- accession_name (required) A unique name for the germplasm line. If your breeding program already has similar lines in the database, please try to follow the same naming conventions to keep the names consistent and to reduce the possibility of duplicate entries.
- species_name (required) The genus and species name of the entry.
- population_name A population is a grouping of accessions stored in the database that can be used by various tools on the website. If you want to add the entry to a population, you can do so here or through the website after it’s been added to the database.
- organization_name(s) The name(s) of the breeding program(s) where the entry originated. Existing breeding program names can be found on the Manage > Breeding Programs page. Multiple breeding programs can be separated by a comma.
- synonym(s) A comma separated list of alternate names for the entry.
- variety(s) For released lines, this can be used to store the released variety name. The variety name should also be listed as a synonym so it can be included in various search functions.
- country_of_origin(s) the entry’s country of origin.
- notes(s) additional comments about the entry.
- accession_number(s) a comma separated list of registered accession numbers (such as GRIN PI numbers). The accession number should include a prefix defining the registration source (such as PI, CItr, etc), followed by a space, then the registration entry number (ex “PI 428501”).
- purdy_pedigree The Purdy pedigree string of the entry. Currently, this is not parsed by the database and will be stored as-is.
- filial_generation The filial generation of the entry.
Add New Accessions
To upload an Accession template:
- Go to the Manage > Accessions page
- Click the Add Accessions Or Upload Accession Info link near the top right corner of the page
- Select the Uploading a File tab
- Click the Choose File button to select your upload template
- Click the Continue button and follow the prompts
View Accessions
Once your upload has been processed, you can find your new Accession(s) by going to the Search > Accessions and Plots page and entering the name of one of your Accessions.
Click the name of your Accession in the results table to open the Accession detail page. Here the Additional Info section should include the property values from your upload template.
Trials 
Use Case: You have one or more trials that were previously designed. You know the plot layout of the trials. How do you add the trials to the database?
There are multiple methods of adding a trial to the database. For this example, we’ll be adding an existing trial by uploading a trial template file (not using the trial design tool). There are two trial template types:
- Single Trial Design
- This trial template includes just the plot information
- The trial information is filled out in a form on the website
- This method can only be used for a single trial at a time
- Multiple Trial Designs
 This is the method we’ll be using in this example
This is the method we’ll be using in this example
- This trial template includes both trial and plot information
- There is no form on the website to fill out when uploading the file
- This method can be used for one or more trials
Trial Properties
The following properties relate to the trial:
- trial_name (required) A unique name for the trial (cannot contain spaces). By convention, this is typically a concatenation of an experiment code, year, and location separated by underscores (ex: “USSRWWN_2008_Blacksburg”).
- breeding_program (required) The name of the Breeding Program that performed the trial. The Breeding Program must already exist in the database (go to the Manage > Breeding Programs page to view and/or add a Breeding Program). For cooperative nurseries, we have created Breeding Programs to contain cooperative nursery trials.
- location (required) The name of the location where the trial was help. The location must already exist in the database (go to the Manage > Locations page to view and/or add a location).
- year (required) The year the trial was held.
- design_type (required) The code for the trial design type. It can be one of:
- CRD Completely Randomized Design
- RCBD Randomized Complete Block Design
- Alpha Alpha Lattice Design
- Augmented Augmented Design
- MAD Modified Augmented Design
- Westcott Westcott Design
- Lattice Lattice Design
- description (required) A description of the trial and any additional notes with relevant information about the trial
- trial_type The name of the trial type. It can be one of:
- Seedling Nursery
- phenotyping_trial
- Advanced Yield Trial
- Preliminary Yield Trial
- Uniform Yield Trial
- Variety Release Trial
- Clonal Evaluation
- genetic_gain_trial
- storage_trial
- heterosis_trial
- health_status_trial
- grafting_trial
- Screen House
- Seed Multiplication
- cross_block_trial
- Specialty Trial
- plot_width plot width (meters)
- plot_length plot length (meters)
- field_size field size (hectares)
- planting_date planting date (YYYY-MM-DD)
- harvest_date harvest date (YYYY-MM-DD)
Plot Properties
The following properties relate to a plot:
- plot_name (required) A unique name (cannot contain spaces and must be unique across the entire database) for the plot. By convention, this is typically a concatenation of the trial name and plot number separated by a dash (ex: “USSRWWN_2008_Blacksburg-PLOT1”)
- accession_name (required) The name of the Accession observed in the plot (must already exist in the database)
- plot_number (required) the plot number
- block_number (required) the block number
- is_a_control use
1for a control/check and0(or blank) for an experimental line - rep_number The replicate number
- range_number The range number
- row_number The row number of the plot in the field
- col_number The column number of the plot in the field
- seedlot_name The name of the seedlot where the planted seed originated (must already exist in the database)
- num_seed_per_plot The number of seed transferred from the seedlot to the plot
- weight_gram_seed_per_plot The weight of seed (g) transferred from the seedlot to the plot
Trial Template
The trial upload template combines the trial properties (which are repeated for each plot) and the plot properties.
| trial_name | breeding_program | location | year | design_type | description | trial_type | plot_width | plot_length | field_size | planting_date | harvest_date | plot_name | accession_name | plot_number | block_number | is_a_control | rep_number | range_number | row_number | col_number | seedlot_name | num_seed_per_plot | weight_gram_seed_per_plot |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AYT_2019_Ithaca | Cornell University | Ithaca, NY | 2019 | RCBD | 2019 Advanced Yield Trial @ Ithaca | Advanced Yield Trial | 1 | 1 | 2019-06-15 | 2019-10-11 | AYT_2019_Ithaca-PLOT101 | VA14W-99 | 101 | 1 | 1 | 1 | 1 | ||||||
| AYT_2019_Ithaca | Cornell University | Ithaca, NY | 2019 | RCBD | 2019 Advanced Yield Trial @ Ithaca | Advanced Yield Trial | 1 | 1 | 2019-06-15 | 2019-10-11 | AYT_2019_Ithaca-PLOT102 | NC14-23372 | 102 | 1 | 1 | 1 | 2 | ||||||
| AYT_2019_Ithaca | Cornell University | Ithaca, NY | 2019 | RCBD | 2019 Advanced Yield Trial @ Ithaca | Advanced Yield Trial | 1 | 1 | 2019-06-15 | 2019-10-11 | AYT_2019_Ithaca-PLOT103 | VA13W-38 | 103 | 1 | 1 | 1 | 3 | ||||||
| AYT_2019_Ithaca | Cornell University | Ithaca, NY | 2019 | RCBD | 2019 Advanced Yield Trial @ Ithaca | Advanced Yield Trial | 1 | 1 | 2019-06-15 | 2019-10-11 | AYT_2019_Ithaca-PLOT104 | NC15-25672 | 104 | 1 | 1 | 1 | 4 | ||||||
| AYT_2019_Ithaca | Cornell University | Ithaca, NY | 2019 | RCBD | 2019 Advanced Yield Trial @ Ithaca | Advanced Yield Trial | 1 | 1 | 2019-06-15 | 2019-10-11 | AYT_2019_Ithaca-PLOT105 | NC14-23373 | 105 | 1 | 1 | 1 | 5 |
Add Trials
To add the trial(s) to the database:
- Go to the Manage > Field Trials page
- Click the Upload Existing Trial(s) button near the top right of the page
- On Step 2, select the Multiple Trial Designs tab
- Click the Choose File button to select your upload template
- Click the Upload Trial Designs button and follow the prompts
View Trial
You can view the trial(s) you added by finding them in the table on the Search > Field Trials page (you can filter the table by entering the trial name in the search box in the top right corner of the table).
You can open the trial detail page by clicking on the name of the trial in the table. From this page you can check the properties of the trial. The Field Layout Tools and Phenotype Heatmap section contains a generated field layout with the plot positions. The Experimental Design section contains information on the field design and the accessions used in the trial.
Traits 
Use Case: You have a list of traits that were observed in your trials. Are these traits available in the database? How do you find the traits? What do you call the traits when uploading data?
Breedbase Trait Management
Breedbase manages traits differently than we did with the original version of T3. Trait management may seem more complicated, but this allows us to more accurately and more precisely define the traits. In addition, trait observations can now be more easily compared to data stored in other databases that use the same trait definitions.
On the original T3, there was a T3 trait dictionary that was a list of trait names with descriptions that defined each individual trait. Trait observations were added to the database by linking each trait value with the T3 trait name.
On breedbase, traits are organized in a trait ontology, which is a controlled vocabulary of trait terms and definitions. The ontology is managed in a collaborative effort through The Generation Challenge Programme’s Crop Ontology project and is made publicly available online on their website. T3 has contributed trait definitions for all three of our crops:
The trait ontology is organized in a tree-like hierarchical structure. Under the root of the tree are trait categories - these are broad categories for grouping similar traits (such as Agronomic, Biotic Stress, Quality, etc). Directly under the trait categories are ontology trait terms, where a trait (in ontology terms) is the entity that is being measured (such as grain yield) but does not include any information about how it is measured or with what units. Directly under the trait is one or more ontology variable terms. The variable term contains information for a single trait as well as information about the method in which the trait was measured and the units/scale the value is recorded in. Observations made from a phenotyping trial must always be associated with a trait ontology variable.
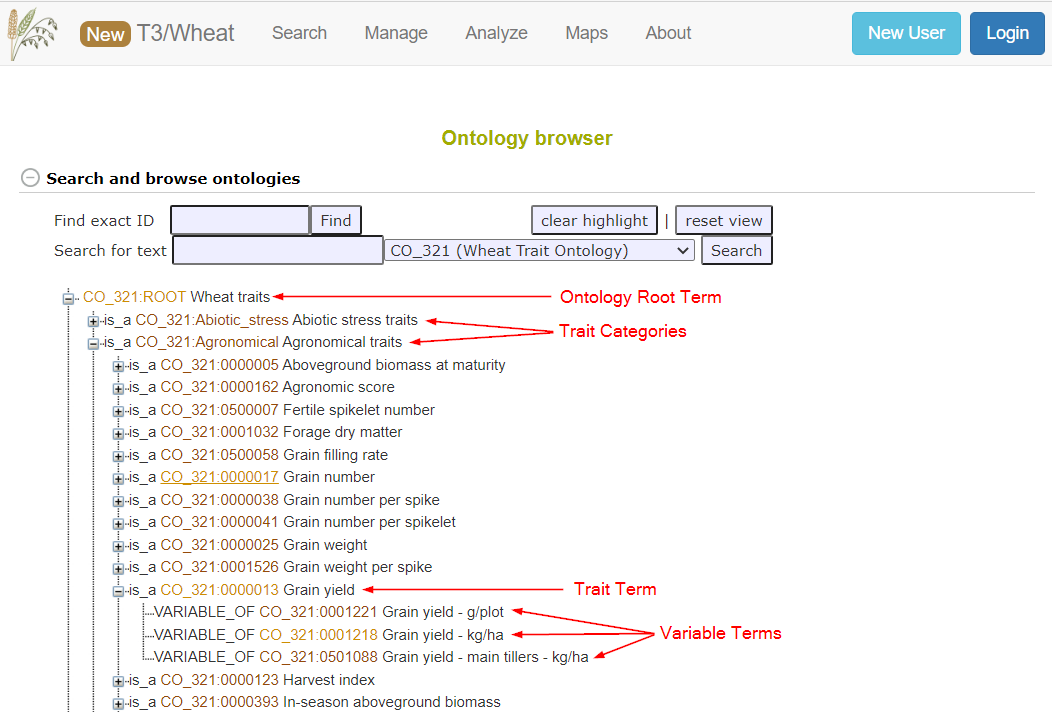
View Traits
To view the traits in the trait ontology, open the ontology browser by going to the Manage > Trait Ontology Browser page. This page will display all of the ontologies loaded into the database - the trait ontology will be labeled Wheat traits, Barley Traits, or Oat traits. Direct links to the trait ontology for each crop are:
Here you can navigate through the ontology tree to find the ontology variable that matches your trait.
Search Traits
Traits can also be searched by name by going to the Search > Traits page. Make sure the trait ontology is selected in the search settings and enter a term to search for.
From the results table, you can choose a term that has a type set to VARIABLE_OF to associate with your trait data.
Trait Usage
When loading your phenotype observations you’ll need to know both the trait name and trait id of the trait variable that matches your observed trait. For example, for wheat grain yield in kg/ha you’ll need to know:
- the trait name: “Grain yield - kg/ha”
- the trait id: “CO_321:0001218”
Trait Lookup Tables
If you’re familiar with the old trait names used on T3/Classic, you can use the lookup tables that link the old trait names to their corresponding breedbase trait variable names and IDs. There is a trait lookup table available for each crop:
Requesting New Traits
If there is no corresponding trait variable that matches your observed trait, you can request to have a new trait added to the ontology. You can either contact us directly or use the trait request form.
If your trait only differs in the units/scale that was measured, we ask that you convert your values to match the scale of an existing trait variable.
Phenotypes 
Use Case: You have phenotype observations from one or more trials that have already been added to the database. How do you add the observations to the trials?
We’ll be adding phenotype observations to the new trials we created by creating a phenotype upload template. We’ll be creating the template manually, but a blank template with the proper trait column headers can be created by the website (from the trial detail page) if you have the traits you observed already in a list.
Create Phenotype Upload Template
We’ll be using the simple phenotyping spreadsheet format. This format has one required column for the plot name followed by a column for each observed trait. The column headers are:
- observationunit_name for the plot name - which are the names we gave each of the plots earlier in the trial upload template.
- trait_name|trait_id is the format used for the column header for each observed trait. For example, for wheat grain yield in kg/ha, the column header would be “Grain yield - kg/ha|CO_321:0001218”. This format can be repeated across multiple columns for multiple observed traits.
Below is an excerpt of the trait observations from the demo dataset:
| observationunit_name | Grain yield - kg/ha|CO_321:0001218 | Grain test weight - g/l|CO_321:0001210 | FHB incidence - %|CO_321:0001149 | FHB severity - %|CO_321:0001440 | FHB DON content - ppm|CO_321:0001154 |
|---|---|---|---|---|---|
| AYT_2019_Ithaca-PLOT101 | 1984.58 | 739.01 | 53.15 | 8.91 | 1.13 |
| AYT_2019_Ithaca-PLOT102 | 2044.19 | 756.15 | 35.59 | 8.08 | 1.75 |
| AYT_2019_Ithaca-PLOT103 | 2356.95 | 787.56 | 36.02 | 20.78 | 1.52 |
| AYT_2019_Ithaca-PLOT104 | 2061.00 | 752.54 | 69.92 | 20.64 | 2.25 |
| AYT_2019_Ithaca-PLOT105 | 1535.94 | 769.60 | 78.59 | 9.40 | 1.38 |
 Multiple trials can be uploaded at once with the same phenotype upload template.
Multiple trials can be uploaded at once with the same phenotype upload template.
Upload Phenotype Template
To upload the phenotype template:
- Go to the Manage > Phenotyping Results page
- Click the Upload Spreadsheet link near the top right corner of the page
- Select Simple for the Spreadsheet Format
- Click the Choose File button to select your template
- Click the Verify button to check the format of the template
- Click the Store button to store the data in the database
View Phenotype Data
You can view the trait observations from the trial detail page (find your trial in the Search > Field Trials page and click the trial name to get to the trial detail page).
In the Phenotype Summary Statistics section you can view a table of trait means and summary statistics and a histogram of trait observations for the trial. Here you can also download a table of all of the trait observations for the trial.
Day Two
Discussion of Day One
Common Problems
- Database URLs
- breedbase.org is a demo website setup by the breedbase Development team which T3 does not have access to
- Links to all T3 Databases are available from https://triticeaetoolbox.org
- Make sure to use the sandbox version for uploading data and the production version for accessing data
- Account Creation/Verification
- most people seem to have received the confirmation email
- a handful of users did not receive the email
- we may need to rethink our email setup
- Excel files
- file names cannot contain spaces
- file type must be the old Excel format (.xls extension - not .xlsx) = Excel 1997-2003 Workbook
- changes we can make:
- make it clear that spaces are not allowed in the file name
- support spaces in file names
- support the “new” Excel format
- Date Formats
- in the trial template: planting and harvest dates have to be formatted as “YYYY-MM-DD”
- Excel has an annoying habit of converting any value it thinks is a date to an Excel-specific representation of the date (an integer)
- to prevent Excel from changing the values: change the column cell type to “text”
- Template Upload Errors
- the “working” dialog would “hang”
- there was either a problem with the template and the error is not reported back to the user
- the upload did not proceed for an unknown reason
- the “working” dialog would disappear with no feedback to the user
- this often due to a problem with the template but it is not reported back to the user
- workaround for now:
- try refreshing the page and try uploading the file a second time
- if it still fails, let us know and we can take a closer look
- changes we can make:
- improve error detection in the files
- more clearly report the problems back to the user
- the “working” dialog would “hang”
Submitting Trials
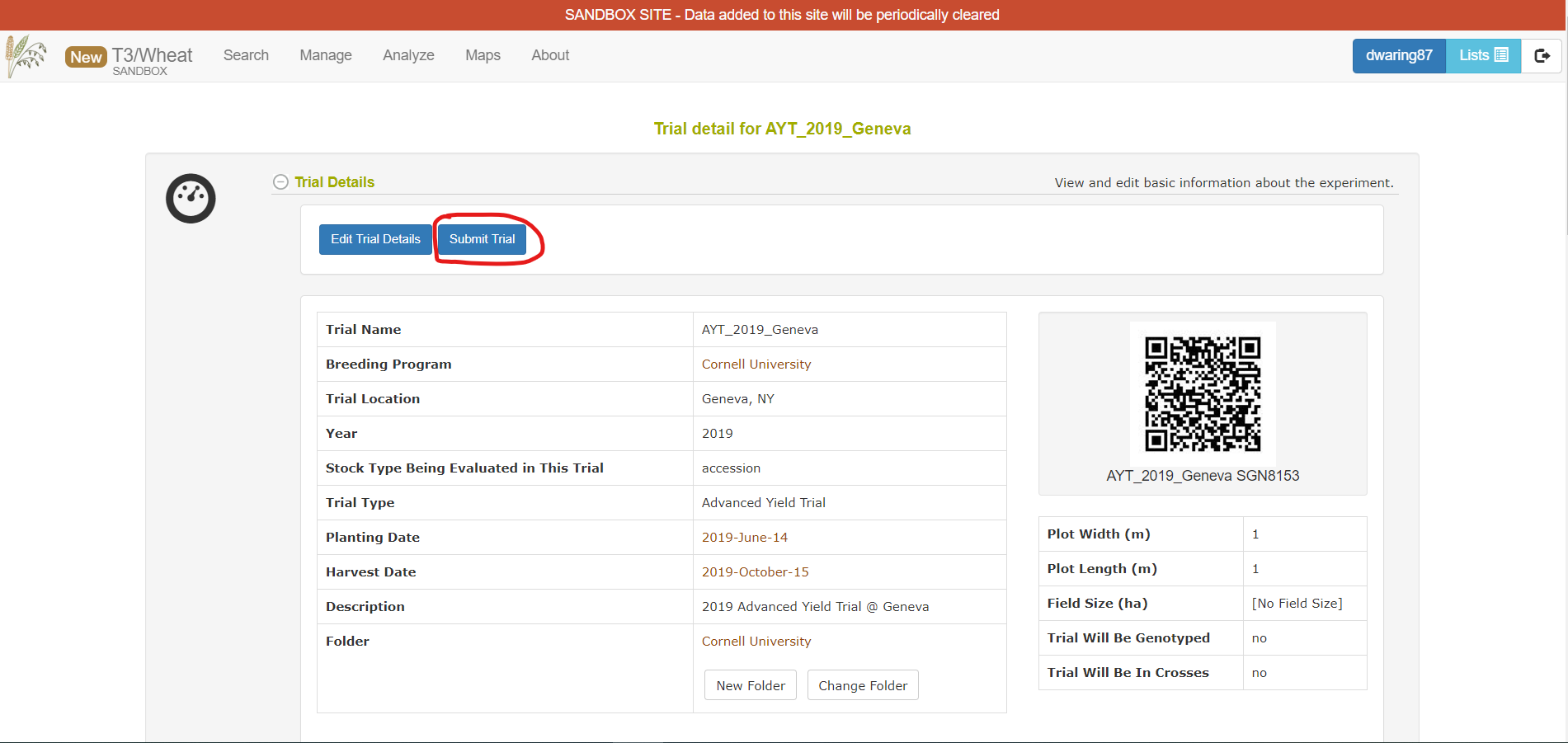
Once you have your trial(s) up on the sandbox, you can submit them to be included on the production database.
From the trial detail page, there is a Submit Trial button near the top of the page. This will tell us that the trial is ready for submission. We will review it and add it to the production database.
Available Shortcuts
Website Tools
There are tools available on the website that can help generate some of the templates we created manually.
- Trial Design Tool This tool can be used to interactively generate one or more field designs
- Phenotype Spreadsheets If you have the traits you are observing in a field trial in a list, you can easily generate a blank phenotype observation upload template with the trait column headers filled in.
- Create a list of traits
- Go to Manage > Phenotyping Results page
- Click the Create Spreadsheet link near the top right of the page
- Select your trials and traits
- Download spreadsheet
Android Fieldbook
Breedbase is tightly integrated with the Android Fieldbook app which can be used to collect phenotype data on an Android tablet or phone directly in the field. The website can be used to create the field layout files that are used to load your field trial into the app. The data files created by the app can then be loaded directly into Breedbase to store the phenotype observations.
Breedbase R Package
If you’re using R to interact with your data, you can use the Breedbase R package to generate the upload templates.
Analysis Tools Overview
Trial Summary Tool
This is a tool that was available on T3/Classic and ported over to breedbase. It can be used to generate LSMeans and LSDs for one or more traits across one or more trials. It is accessible from the Analyze > Summarize Trials menu.
To use the tool, you will first need to have the trials you are interested in summarizing in a list.
Trial Analysis Tools
These tools are included with breedbase and are available in the Analysis Tools section of a trial detail page.
- Correlation
- ANOVA
- PCA
- Clustering
- Heritability
Breedbase Analysis Tools
These tools are built in to breedbase and could be made available.
- Selection Index
- Genomic Selection
- GWAS
Some of these tools are not yet available because they require some work from us to get them up and running. If you are interested in using any of these tools, please reach out to us and let us know. This will allow us to prioritize setting up tools that we know people are interested in using.